NF-kB
Click here to
gain access to an excellent review. You may recognise a few of the diagrams!!
Nuclear Factor necessary for Ig kappa (k) light chain transcription in B cells.
Referred to a the prototype of dimeric
transcription factors
OVERVIEW
(adapted from Baeurele et al., 1996, Cell 87,
13-20)
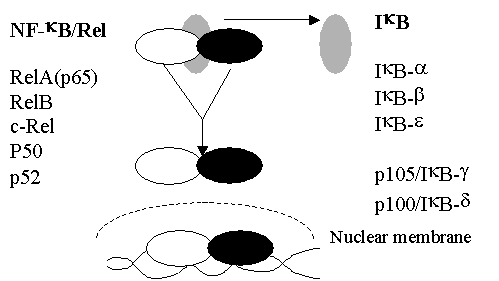
NF-kB exists as a dimer in the cytoplasm – complexed
to an inhibitor protein
IkB that prevents trafficking to the nucleus.
NF-kB activation is associated with the release of
IkB – allowing nuclear targeting.
NF-kB
forms
·
p50
(NF-kB1)
·
p52
(NF-kB2)
·
p65
(RelA)
·
cRel
·
RelB
STRUCTURE
Related to the
proto-oncogene , c-Rel.
“Rel” proteins were first characterized
as cryptic DNA binding proteins
Formed from two DNA binding
subunits made up from homo- or
heterodimers
“rel” motif proteins. This
is approximately 300a.a.
The Rel area
contains
·
Dimerisation
motifs
·
DNA
binding region
·
A
nuclear localisation signal.
·
Interaction
with the inhibitor protein - IkB (see later notes).
The
C –terminus (not found in p50 or p52) contains varies greatly in each rel
protein
and is responsible for transactivation.
Interaction with
DNA
Determined for the human and mouse p50:p50 homodimer
– (see Ghosh et al., 1995, Nature, 373 : 303-310 in General Office Papers)
– though the most common interaction is p50/p65.
· Resembles a “butterfly”- with two pairs of wings connected to a central
core of DNA.
· The Rel region folds into two domains- connected by a 10 residue linker
(238-247aa)
· C-terminal domain (248-350aa) – core b-sheets – similar in structure to
the immuno-
· globin family.
· This region is responsible for dimer formation- which brings together 10
loops (five from
· each subunit) – to fill the major groove of a DNA molecule and generate
a sequence
· specific DNA-binding surface.
· Effectively wraps around the DNA molecule.
· Binding is augmented by the a-helix in the N-terminal region which bind
to the minor grove.
· Any variation in this dimer/loop interface will affect NF-kB specificity
Genes regulated by NF-kB have “ kB” sites of the following consensus DNA sequence
5’ GGGRNNYYCC 3’. R
= Purine, Y = Pyrimidine, N = any base.
(the variation reflects the varying combinations of
homo and heterodimers
that
can form.)
The sequence results
in an unusual conformation of DNA-
One turn of DNA molecule
usually consists of 20bp but is 10.7bp in this region-
This tight twist
results in a deep major groove.
The Interaction with IkB
(Ghosh et al., 1995, Nature, 373 : 303-310)
Nuclear localization signal is found at
residues 360-364 on NF-kB, which is masked by IkB.
Found just above the dimer interface- in the
flexible linker domain. - therefore the interaction
with IkB is likely to alter the “butterfly”
configuration.
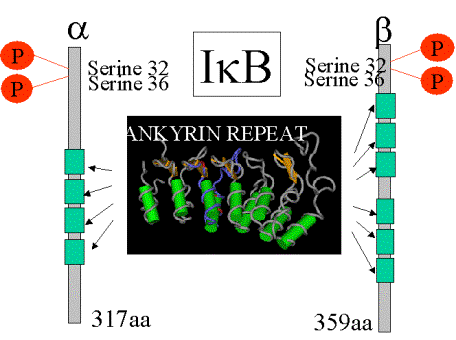
I-kB: The NF-kB
sequestrator
Bind
to rel region of NF-kB to block the nuclear localization signal so that the
transcription factor remains in the
cytoplasm.
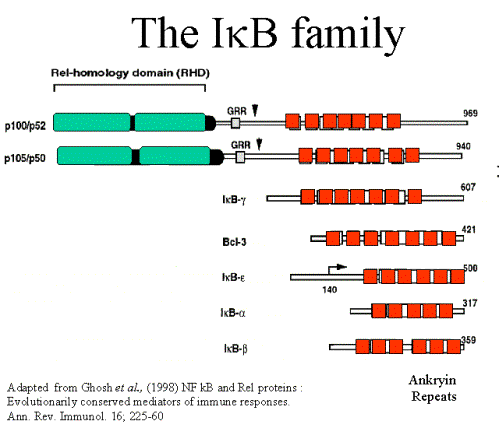
IkB inhibitors all
have 5-7 ankyrin (30aa) repeats domains
Release
·
Involves
the phosphorylation of serines 30 + 32 (N-terminus).
·
Followed
by ubiqutinisation of lysines 21 and 22 and degradation by the 26S
proteosome. C-terminus is essential for the interaction with the “proteosome”
–
as
part of ubiquitin – mediated degradation.
What is ubiqutinisation?
Protein-degradation
service – proteins are targeted for proteolytic digestion by
being
decorated with ubiquitin
·
E2
“ubiquitin conjugating enzyme”. This may directly transfer ubiquitin to
lysines on the target protein followed by poly-ubiquitinisation. Further ubiquitin
proteins are added to lysines lying within the
conjugated ubiquitin protein.
·
Proteosome -the polyubiqutin chain
targets the protein for degradation.
Click for more information on ubiquitinization
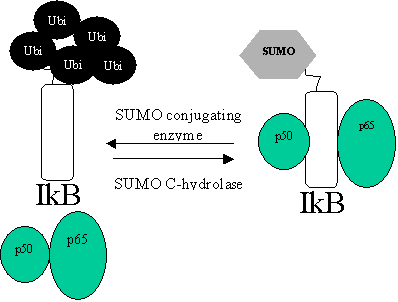
Suppressing IkB degradation: Protection using SUMO-1.
SUMO-1
binds to the same lysines used for ubiquitinisation. This creates a “privileged
pool of IkB” that doesn’t respond to cytokine
signals.
Forms
of IkB
Four
main forms of IkB with differing affinities for NF-kB forms - suggesting the
possibility
of selective degradation to release certain NF-kB forms.
IkB-a: 37kDa protein. The “classical IkB” in terms of regulation.
IkB-b: 45kDa protein. With IkBa is responsible for inhibiting the majority
of p50:p65.
IkB-e: Only associated with RelA and
c-Rel – indeed relA is seldom complexed
with
other forms. Only slowly degraded following stimulation. Regulates specific
genes e.g. IL-8.
IkB-g: 70 kDa protein only detected in lymphoid cells. Identical to the
c-terminal
regions
of p105. Probably only inhibits p50 and p65 homodimer.
Viruses: African
swine fever virus – produces an IkB –like protein (A2302) which
can suppress NF-kB activation and therefore the
inflammatory response.
A REGULATED NEGATIVE FEEDBACK LOOP
·
NF-kB
induces the expression of many of suppessors (IkB).
·
IkB
have nuclear localization signals -so
can remove NF-kB from kB sites and suppress expression.
·
Cells
must avoid NF-kB effects being transitory but retain this as a potential
negative feedback mechanism
·
This
is done to a great degree IkB-b.
·
Both
phosphorylated and non-phosphorylated forms of IkBb bind to NF-kB but only the
phosphorylated form can prevent DNA binding ie. remove
the transcription factor.
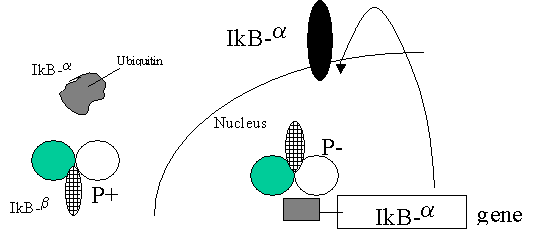
STEP1
Thus, IkB-a is ubiquitinated and degraded –
STEP2.
NF-kB moves to the nucleus
STEP3 Can
bind to IkB-a promoter which has NLS which can remove NF-kB
from kB sites
STEP4 BUT
this can be blocked if NF-kB is bound by
non-phosphorylated IkB-b -( Persistent activation)
STEP5.
Phosphorylation of IkB-b will lead to this
removing NF-kB from sites (Suppression of gene expression)
Biogenesis of NF-kB - p50/p52.
p50
and p52 are synthesized from the N-terminus of larger precursors p105 and p100
respectively.
At C-terminus – “ankyrin repeats” – these allow
protein / protein interactions.
These
can interact with the NLS to suppress nuclear trafficking –as does IkB
p50/p52 proteolytic cleavage is ATP dependent
and involves ubiquitinisation
Cleavage of the ankyrin repeat regions immediately results in nuclear trafficking.
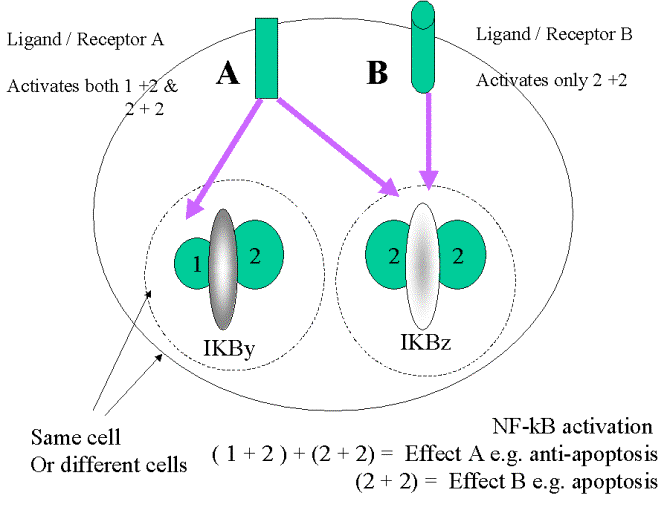
NF-kB multi-tasking
The variable interactions between NF-kB
monomers with varying forms of IkB allow a similar
mechanism to be used for very differing
effects.
Evidence: -
(i) Knock-outs
Rel
A-/- Causes apoptosis of liver cells in
mouse prior to birth
(i.e. primarily
anti-apoptotic)
Rel
B-/- Lethal multiple organ inflammation.
(i.e. primarily
anti-inflammatory)
c-Rel-/-
B and T cell
deficiencies
p52-/- Altered lymph mode
architecture
(ii) “Artificial” formation of dimers has different effects.
p50/c-Rel, p65 (Rel-A) /p65 (Rel-A), p65
(Rel-A)/c-Rel dimers will activate transcription
p50 & p52 homodimers will suppress
transcription.
How do cells influence what dimers will be form/ be activated?
(i) Tissue specific expression of subunits .
This influences which homo-/heterodimer will
predominate.
(ii) Variable interactions with IkB - forms
(iii) Variable upstream activation –
influencing the phosphorylational
changes