Major pathogenicity functions are encoded by hrp genes
In the beginning… it was observed that
biotrophic
bacteria…
1.
tended to be
in contact with plant cell wall.
2.
must be
metabolically active to elicit disease
(or
resistance).
In early 1980s- transposon tagging resulted in mutants that were unable to elicit a HR or acts a pathogen (hypersensitive response and pathogenicity).
The hrp- phenotype
Besides the above
· hrp mutants were unable to grow in
planta
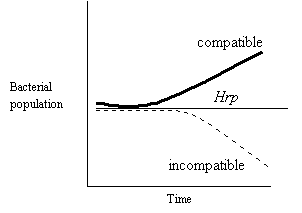
· but it is not required for growth on minimal
media
plates – therefore they are
not an auxotrophic mutant.
· Certain
defence genes are still induced – phytoalexins
and PR proteins- an ancestral resistance mechanism?
Genetic organisation
It was noted that hrp
functions were clustered.
P.
s. pv. phaseolicola 22kb
in eight transcriptional units
P.s. pv. syringae 25 eight
X.c. pv. campestris
23 six
Ralstonia solanacearum 25 seven
It
was noted that avirulence genes
were often linked to these
loci.
Note: hrp genes are
not avirulence genes since isolated hrp genes
cannot elicit the
hypersensitive response.
BUT – hrp + avirulence genes (on single clone- pHIRII)
can elicit symptoms when introduced into a non-pathogen e.g. Pseudomonas
fluorescens or E. coli.
Biochemical
function
Database searches revealed
that many hrp genes exhibited
homology to Type III
secretion systems.
Type II
Sec
–dependent pathway – involving a
two step mechanism.
· Export
to the periplasm. ATP-dependent.
Followed
by cleavage of a N-terminal “segment”
by a
signal peptidase.
· Passive
export out of the outer membrane.
Type I
Sec-independent.
A one step (ATP-dependent) mechanism
through a channel or a gated
pore.
Type III
A sec-independent
two-step mechanism. Commonly used
to deliver proteins from the
bacterial cytoplasm to the
eukaryotic cytostol.
Key features
· Absence
of signal peptide as used in sec pathway
· Requirement
for host cell contact.
Hrp
genes can be split into the following functional groups –
hrp A → outer membrane proteins → forms pilus-ike
structure.
One → outer membrane associated
lipoprotein
(hrp
C.)
Five
→ inner membrane associated proteins
(hrp RSTVJ)
Two → cytoplasmic proteins
(one
is an ATPase, hrp N).
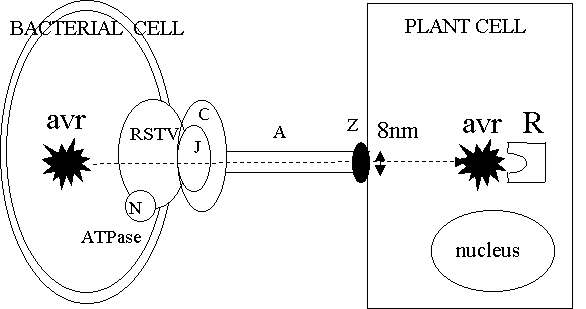
Seven of the nine hrp proteins exhibit homology to
other Type III particularly
that found in Yersinia-
designated HR
and conserved = hrc
Yersinia – Plague and gastroenteritis
Shigella -
Dysentery
Salmonella
– Food
poisoning
E.coli - Diarrhoea
However : these pathosystems end with bacterial entry into the animal cell.
This is NOT the case with
plant bacterial pathogens.
Hrp Gene Regulation
hrp gene
expression influenced by
Carbon/nitrogen source, pH,
osmolarity, temperature and
possibly plant signal molecules.
Gene expression is dependent
on hrpR and L products detecting
the metabolic status of the
cell (“sated” and “hungry”) and interacting
with sigma 54 to bind to a
regulatory region (the hrp box) and thereby
activating hrp gene
transcription.
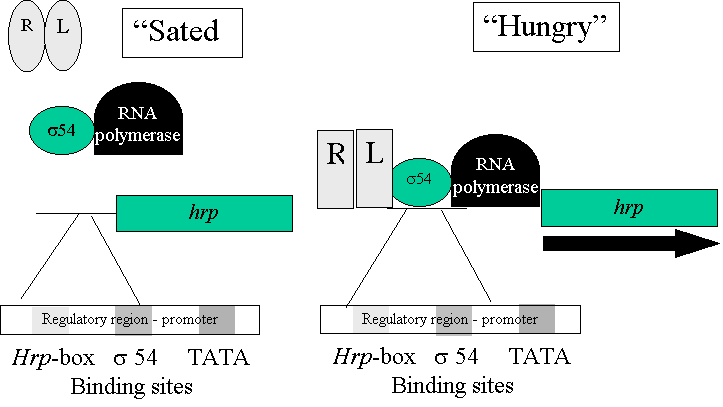
Hrp box – GGAACCNA – N14 - CCACNNA
What is delivered?
Hrp –system is NOT required for
soft-rot enzymes in
Erwinia
carotovora or toxin genes in Pseudomonas
syringae or
EPS genes in Ralstonia solanacearum!
What is delivered in analogous systems?
Yersinia – YopE/H : Interacts
with actin cytoskeleton to
suppress phagocytosis.
Salmonella – SipA : Induces membrane
ruffling by inhibiting
F-actin depolymerisation.
P. aeruginosa – ExoS :
targets small GTP binding protein
exchange
factor.
Surprisingly : “Avirulence” gene products
are delivered.
So what are the virulence
functions of these proteins?
· AvrBs2 exhibits homology to
agrocinopine
synthase in Agrobacterium.
Directs transformed
cells to produce a new carbon source- “cellular
metabolic reprogramming”?
· AvrBs3 localised to the nucleus where it may
activate the transcription of unknown genes.