Apoptosis
For excellent general reviews on apoptosis click
here.
and here.
The most heavily investigated PCD death-mechanism.
Derived from a Greek term describing autumn
leaf-fall.
Animal cells use cell-death :
(1)
Developmentally e.g. finger separation during
embryogenesis
(2)
control organ development and
size e.g. skin.
inappropriate activation leads to e.g. AIDS
inappropriate inhibition leads to cancer.
(3)
destruction of infected cells
BUT
cell contents are inflammatory so cannot simply rupture!
Click for apoptosis movies : Movie 1  and
Movie 2
and
Movie 2 
Hence apoptosis involves
(i) nuclear
and cytoplasmic condensation margination of
chromatin along
nuclear envelope
(ii) membrane blebbing compartmentalisation of
nuclear and
cytoplasmic material
→ apoptotic bodies
(iii)
DNA ladder formation
DNA fragmentation into
200bp increments corresponding
to internucleosomal
spacing.
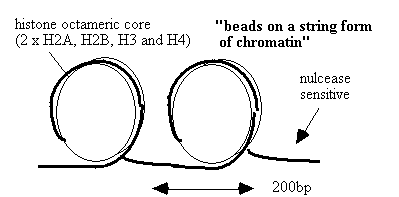
Used as a diagnostic marker for
apoptosis.
Caspases : The workhorses of apoptosis
CYSTEINE-PROTEINASES
a. k. a Caspases.
named as activity is dependent on cysteine in active site
Proteinases which will cleave at aspartate residues within proteins

Translated as inactive precursor polypeptide
separated by aspartate residues
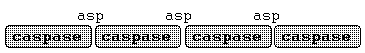
represents a positive feedback mechanism
SUBSTRATES
(A) AFFECTING CELL
SHAPE
(i) Microfilaments : gelsolin, actin and
intermediate filaments.
Alters cell-shape and movement during
apoptosis
(ii) Lamin :
cleavage unpins nuclear envelope
(B) AFFECTING DNA
(iii) Caspase
Activated DNase (CAD)
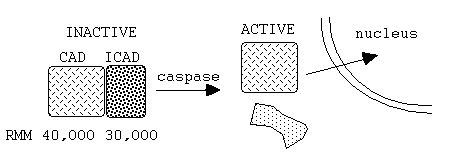
(iv) poly-(ADP-ribose)
polymerase- (PARP) used in DNA
repair . Contains asp-glu-val-asp sites "unhooks" DNA repair from damage
A suppressors of apoptosis
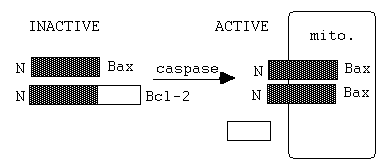
Bcl-2 onco-gene
giving rise to B-cell lymphoma
an
intracellular membrane protein.
Bcl-2 also has a asp-glu-val-asp site-->
therefore a caspase substrate!
Digestion acts to (i) inhibits caspase function
(ii) breakdown product
- Bax, stimulates which forms pores
in mitochondrial membrane.- this releases cytochrome c.
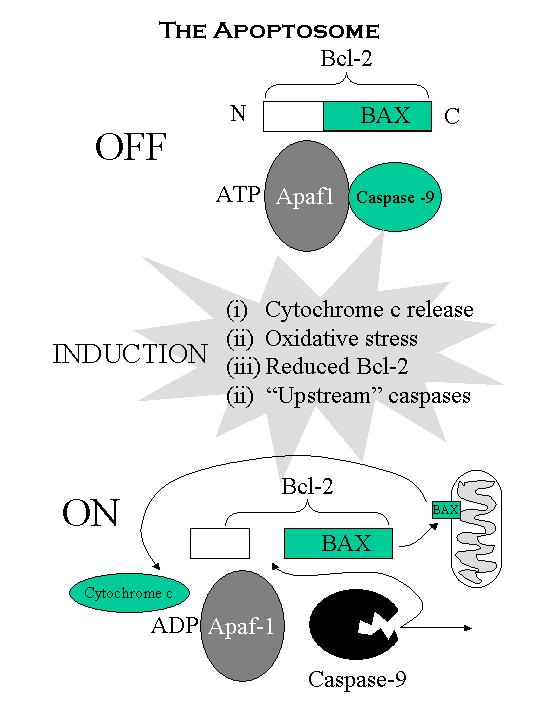
The regulator the apoptosome
How
can Bcl-2 suppress caspases and be a caspase substrate?
Revolves around another complex: the apoptosome
This is a complex formed between Bcl-2/caspase
9/ and Apaf-1
Apaf-1 (i) essential for
cell-death but is not a caspase.
(ii) processes caspase into an active form.
(iii) dependent on ATP hydrolysis ->
conformational
change but is
absolutely dependent on cytochrome c released from the mitochondria.
Bcl-2 binding prevents ATP hydrolysis (and Bcl-2 prevents cytochrome
release)
1
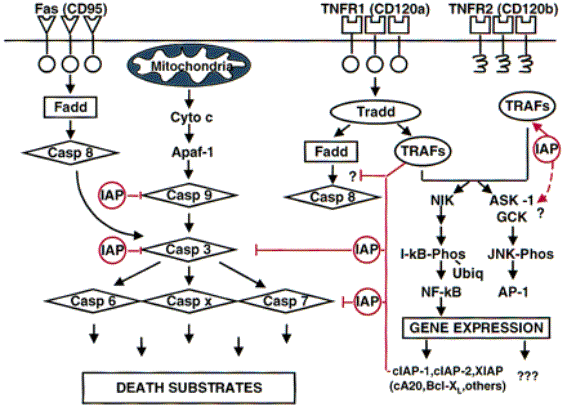
TNF, IL-1 and NF-kB and the
regulation of apoptosis
TNF
and the FAS pathway
TNF-a has well-characterised apoptotic
function by interaction with the FAS-FADD pathway.
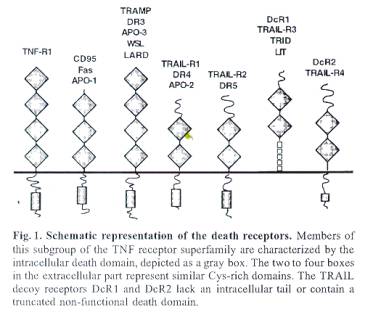
The Fas receptor (CD95) is part of the TNF-receptor
superfamily.
FAS signalling is not suppressed by Bcl-2 thus does not
use the apoptosome mechanism.
FAS-Pathway
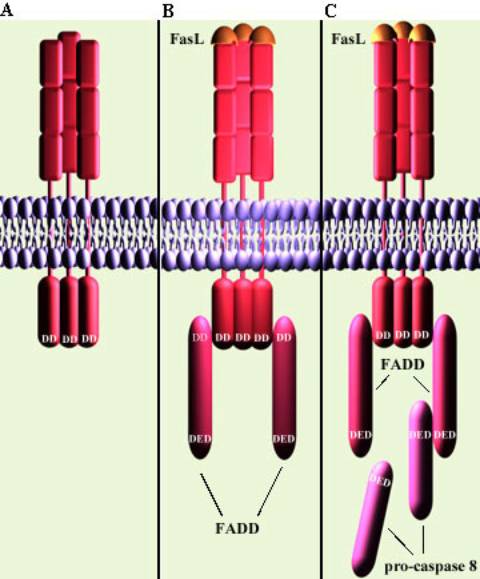
A. Death
receptor Fas with intracellular death domain (DD)
B. Fas Ligand binds to Fas. Receptors
cluster and the Fas death domain associates with
the death domain on the adaptor protein FADD
C. This death-inducing signalling
complex (DISC) recruits pro-caspase 8 (aka FLICE)
molecules via the death effector domain (DED) on the FAD

TNF Pro-apoptotic pathway No 1
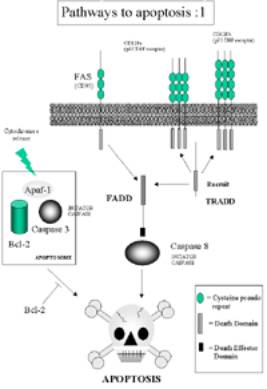 Click here for larger image
Click here for larger image
LI-1 Pro-apoptotic pathway No 2
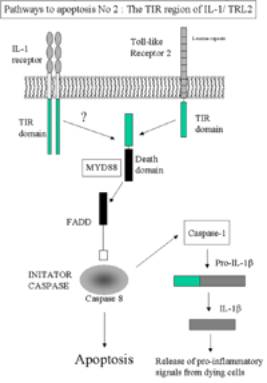 click here for larger image
click here for larger image
It is possible for IL-1 to initiate apoptosis via the
adaptor protein MyD88. This is a has both a TIR domain and death domain
Apoptosis can be intiated via FADD- Caspase 8 mechanism.
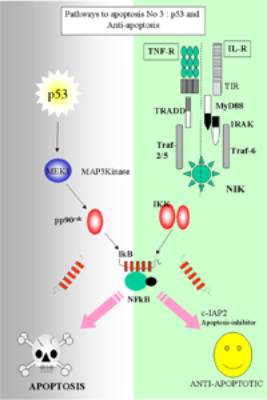
Pro and anti-apoptosis (Pathway No 3)
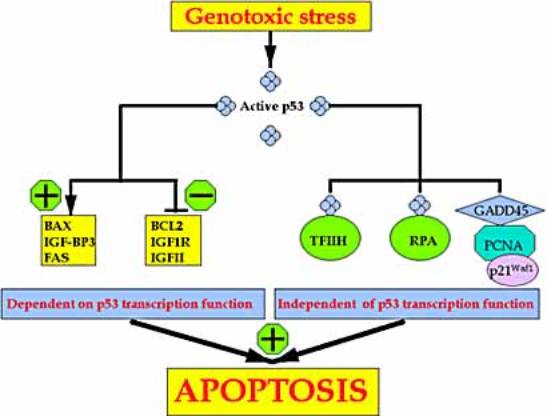
p53
Functional p53 is thought to provide a protective
effect against tumorigenesis,
and indeed, mutations of p53 have been found in
nearly all tumor types and are
estimated to contribute to around 50% of all cancers.
Structural and functional aspects of p53
There are four conserved domains in p53:
1. The N-terminal domain is required for transcriptional transactivation
2. A sequence-specific DNA binding domain
3. A tetramerization domain near the C-terminal end
4. The C-terminal domain interacts directly with single stranded DNA.
Activation
Stress signals (e.g. hypoxia, radiation, DNA
damage or chemotherapeutic drugs ...) activate
·
p53 activation,
·
ubiquitin-dependent degradation of the p53
protein is blocked.
The resulting increase in p53-dependent gene
transcription leads to
·
the p53-mediated induction of programmed cell
death
·
and/or cell cycle arrest.
p53 Target
Genes
·
Wild-type p53 binds to specific genomic sites
with a consensus binding site
5'-PuPuPuC(A/T)(T/A)GPyPyPy-3'.
·
p53 binds as a tetramer and stimulates expression
of downstream genes that
negatively control growth and/or invasion or are mediators of apoptosis.
·
It was predicted that the expression of about
200-300 genes might be controlled
by
p53 transactivation.
p53 and apoptosis.

p53 induces
·
the expression of proteins that target both the
mitochondrial- and the
death-receptor-induced
apoptotic pathways,
·
represses transcription from several death-inhibiting genes.
·
They include the ability of p53 to drive
relocalization of death receptors
such
as Fas/CD95 from the Golgi to the cell surface
·
Murine
double minute 2 (Mdm2), is a transcriptional target of p53.
Mdm2
binds to p53 and targets p53 for ubiquitin/proteasome-dependent
degradation.
Ubiquitination (Ub) of p53 by Mdm2 probably also enhances
the export of p53 from
the nucleus to the cytoplasm, where degradation takes place.
pp90rsk
· forms part of MEK1/2-MAPK (ERK1/2) signaling pathway-
· originally isolated as an Insulin associated kinase
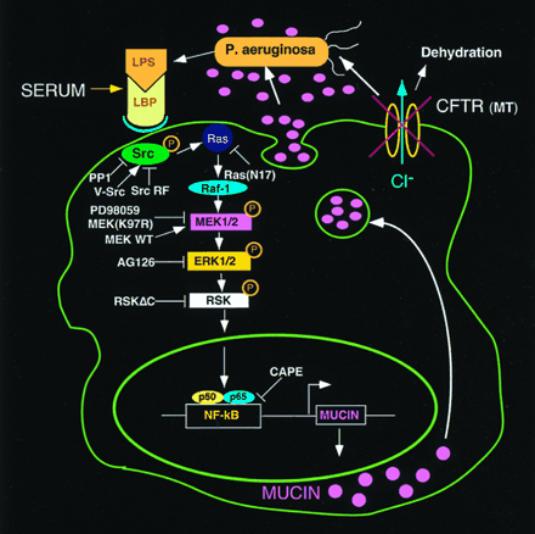
·
Schematic diagram showing steps in the signaling
pathway by which
P.
aeruginosa up-regulates human MUC2 mucin gene
transcription.
·
P. aeruginosa
releases LPS, thereby activating a c-Src-Ras-Raf-1-
MEK1/2-MAPK
(ERK1/2)-pp90rsk pathway leading to the activation
of
NF-![]() B
mediated MUC2 transcription.
B
mediated MUC2 transcription.
·
LBP,
LPS-binding protein. The overproduced mucin, in concert with
abnormal airway lining fluid secondary to CFTR mutation,
leads to airway
mucus obstruction and
lung failure.
 Click here for larger image
Click here for larger image
NF-kB mediated suppression of
apoptosis
Two main
functions
(a)
Inducing the
expression of superoxide dismutase (SOD)
(b)
Inducing
the expression of IAPs inhibitor of apoptosis.
Deveraux and Reed (1999) Gene and Development 13, 239-252.
IAP proteins characterised by having BIR domains
BIR = Baculoviral IAP repeat
Conserved spacing of cysteine residues
(Cx2, Cx6, Wx5, Dx6,
C)
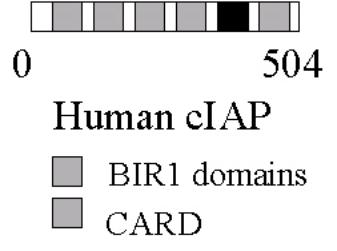
CARD = Caspase recrutiment
domain
IAPs bind to an inhibit a series of CASPASES by bind to the
BIR domains