Plant Viruses: An Overview
Viruses
are nucleic acid protein complexes which multiply in living cells by hijacking
the hosts’ replicative machinery.
One
quarter of all known viruses (>2000) attack plants.
Nomenclature –
from
original plant they were isolated from – and a description of the symptoms
e.g.
Tobacco mosaic virus, Prunus necrotic
ringspot virus.
Tomato spotted wilt virus.
Poor
system since symptoms vary so greatly.
Symptoms:
Plant
viruses do not cause disease through toxin production but disruption of the
cellular processes – hence many viral infections resemble nutrient
deficiencies.
-
Infection with all viruses cause “dwarfing and stunting”
-
Localised chlorotic / necrotic lesions
- Patternings
Mosaic symptoms – arise from 100,000 – 10,000,000 viruses
per cell
Ring-spots
–chlorotic / necrotic rings.
-
Other viruses have no obvious additional symptoms – the latent viruses.
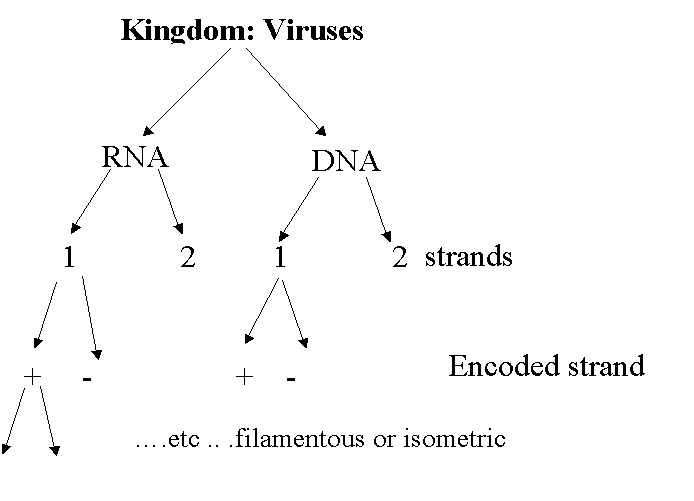
Classification:
Functional Morphology
1.
Shape – coat protein dependent.
-
Rod - (15 x 300nm) “spherical” polyhedral diameter – 17nm in diameter
-
“Bacillus-like”
52-75nm x 300-350nm.
2.
Genome- type and form of nucleic acid
Tobacco Mosaic Virus: A viral paradigm
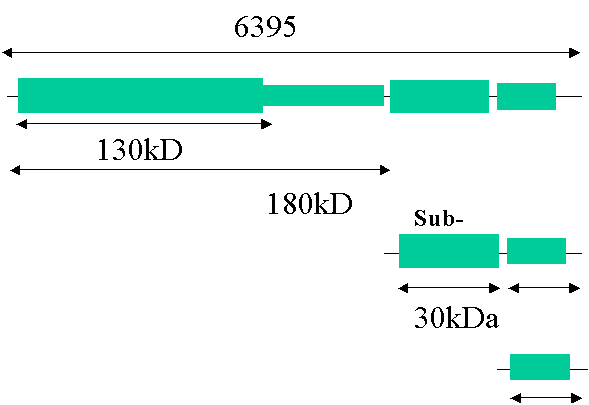 Genome A single RNA stranded (+) virus
Genome A single RNA stranded (+) virus
There
is a leaky STOP codon at the end of the 130KDa gene allowing read through to
form the 180KDa protein.
 Genome
– function
Genome
– function
Note:
Coat protein serves not only a protective role but helps in systemic
dispersal.
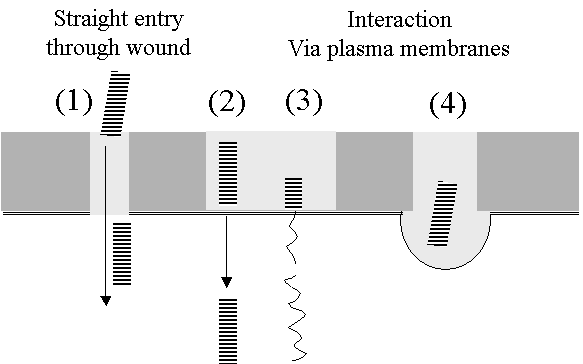
INFECTION PROCESS
Very easily by plant rubbing. Wounding is important -
Virus is very stable. Purified TMV is infectious after 50years storage in a fridge.
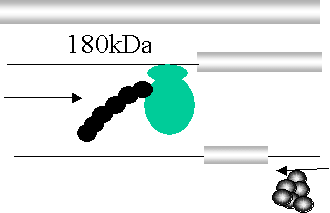
Uncoating
Uncoating
begins 2-3min after infection.
The + RNA is effectively mRNA so can bind ribosomes.
Ribosomes bind to 5’ end and displace coat protein only until the entire 180kDa region is exposed.
The
expressed replicase binds to the 3’ end to uncoat the rest of the virus.
The
process is complete within 30min.
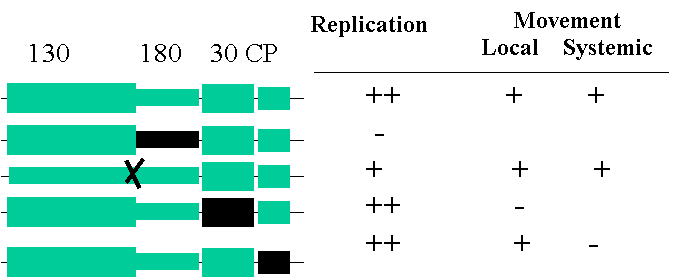
Replication
+ strand is encapsidated, so need negative strand to
for further viral genomes.
Also this will serve as a template for the synthesis of
subgenomic RNAs for MP + CP and CP alone.
This
is necessary since both MP and CP are encoded on the “opposite” – strand.
130kDa
/ 180KDa show homology to RNA dependent RNA polymerase - 180KDa protein is essential for
replication not so the 130KDa.
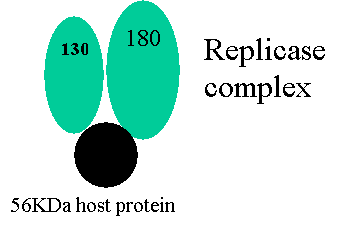
Replicase
complex –
![]()


Replicase
also makes the “incomplete” RNA copies
known as “subgenomics”
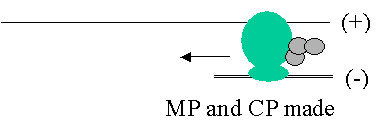
The
use of subgenomics allows simultaneous RNA replication and MP / CP translation
to occur.
Otherwise
this would be attempted on the same molecule!!
Where
is this occurring?
After penetration of the tissue –
(1) the nascent viroids accumulated into “viral factories” –
…in ER where replication predominates
(2) Then disassociate as MP – RNA complexes which are associated within the cytoskeleton.
(3) MP-vRNA accumulates at plasmodesmata.
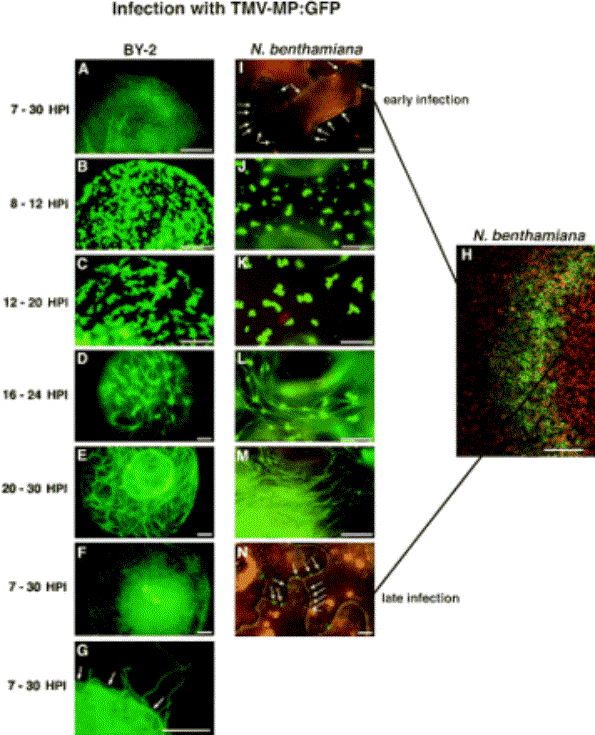
Accumulation and distribution of fluorescent MP:GFP in the population of infected tobacco BY-2 protoplasts and N. benthamiana at given times after inoculation. Note in (B, C, J, K), MP:GFP becomes detectably associated with small irregular fluorescent structures – viral factories in the endoplasmic reticulum. In (D, E, L, M), the irregular fluorescent structures decrease in size and appear to be interlinked with filaments (microtubules). Finally, (G, N) fluorescence becomes associated with the plasma membrane (plasmodesmata). Bars = 10 µm.
Movement
Movement
is governed by source - sink relationships. Hence symptom tend to appear in the
youngest leaves
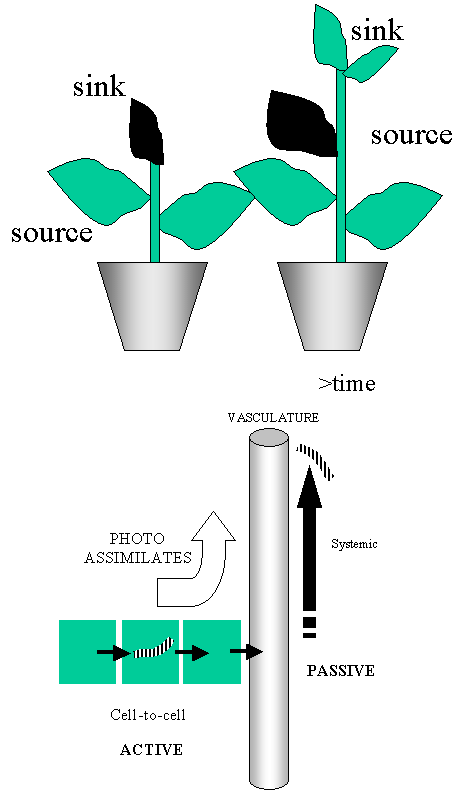
Cell-to-cell (influx + efflux) using
plasmodesmata.
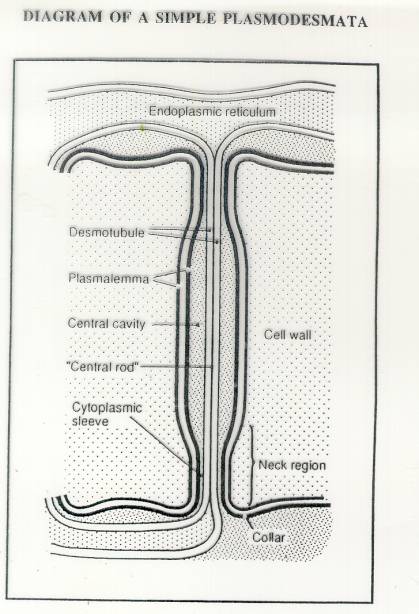
These
are “symplastic” connections between plant cells.
The center of the plasmodesmata is termed the desmotubule and is made
up of
compressed endoplasmic reticulum. The region between the
desmotubule and the plasma membrane is the
cytoplasmic sleeve, the major conduit through which molecules pass
from cell to cell. Electron dense
substructures line the cytoplasmic sleeve between the DT and the
plasma membrane.
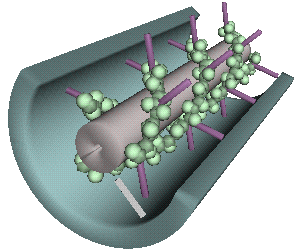
Both globular particles and
elongated spokes appear to interconnect these two membranes and may act to expand or contract the cytoplasmic
space to increase or restrict transport. Cross-sectional views reveal that PD are subdivided into 2.5-nm diameter
microchannels
Plasmodesmata
allow the movement of 1mm 8 to 10 cells
per day in leaf parenchyma.
Cell-to-cell
movement (influx + efflux) aided
by the 30KDa (P30) “movement
protein” which enlarges plasmodesmatal channels.
Four
main functions
(i)
Binds
TMV RNA
(ii)
Interacts
with the cytoskeleton to facilitate transport
(iii)
>
the “pore size” of the plasmodesmata
(iv)
Interacts
with a cell wall receptor
Movement of MP-Nucleic acid complexes
Stage 1 – Nucleic acid become coated with MP
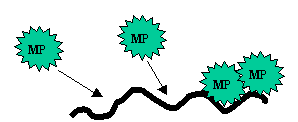
The
particle size is now 2-2.5nm
P30
binding to DNA is not sequence specific.
·
In vitro P30 can bind to any RNA molecule –
·
MPs
from other viruses can substitute for the TMV MP and move TMV RNA to the
plasmodesmata.
Stage 2: The RNA-MP interact with the cytoskeleton
·
On
microtubules – composed of tubulin
·
On
microfilaments – composed of F-actin.
These are the “superhighway” by which the MP-RNA is
targeted to the plasmodesmata.
Stage
3: Interaction with the plasmodesmata.
Plasmodesmata
have a usual pore size of 1.5cm
Exclusion limit is 0.75- 1.00 KDa without MP
>10KDa with MP
Stage 4: Interact with cell wall receptor
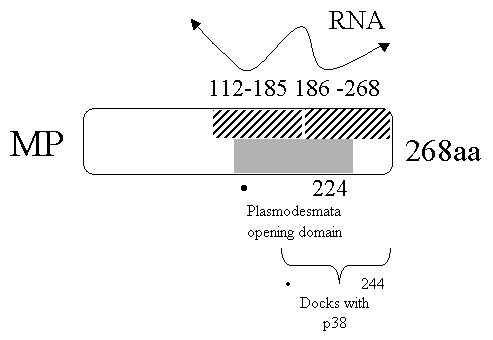 Once
reaches the cell wall/plasmodesmata interacts with a p38 protein.
Once
reaches the cell wall/plasmodesmata interacts with a p38 protein.
P38
appears to mediate RNA / and/or protein trafficking.
One
working model predicts the following.
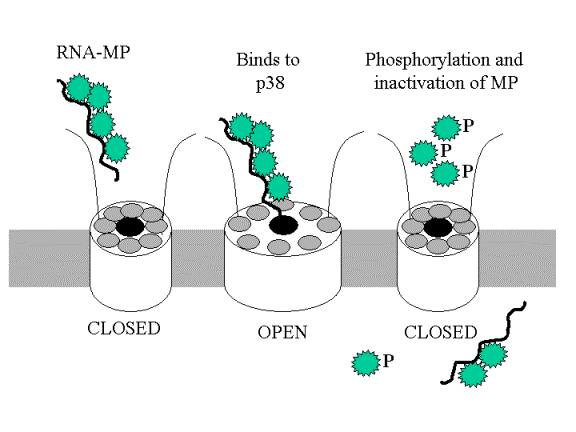
Note, phosphorylation of MP stops their interaction with P38 and causes their partial
release from the viral RNA - but also
the movement of virus to the next cell.
Have virus hitched onto a mechanism
that plants use to control gene expression?
The “Knotted” story.
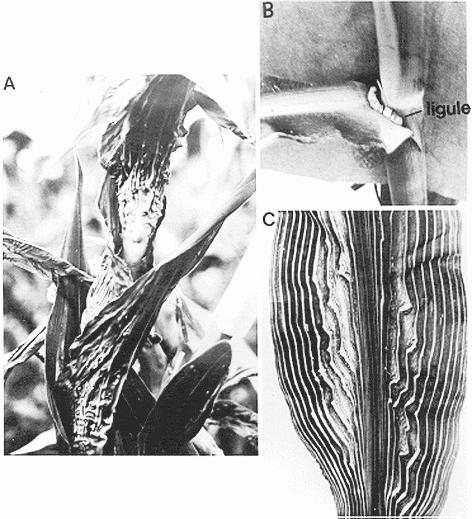
The knotted1
mutation was first isolated as a dominant mutation in maize and produced a “knotted” leaf phenotype
(see above).
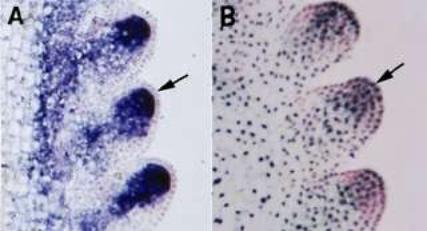
Comparison of the patterns of KN1 mRNA expression and protein accumulation shows that they do not coincide. While expression of the mRNA is excluded from the outer cells in the embryo (see A above) and shoot meristem, but that is where the KN1 protein is localised (see B above) !!
It has been shown that KN1 movement from the site of
transcription to protein accumulation is regulated by plasmodesmata
i.e. that have important developmental role.
Could the
virus MP-RNA interaction with
plasmadesmata be exploiting this function?
Systemic Movement Entry into phloem –Entry into the phloem
(i)
In
the phloem companion cell – virus disassembles
(ii)
A
CP-MP-viral complex is this formed to enter sieve element.
(iii)
In
the phloem CP and vRNA form viral assembly complex.